

If you’ve read through the olfactory pathway page and are interested in a more detailed description of how odorants bind to olfactory receptors (ORs), look no further!
A ligand is a chemical compound or protein that is capable of binding to a protein. The ligands we care about in olfaction are odorant molecules. Molecular interactions, steric effects (energy-driven interactions that influence a molecule's shape and reactivity), and other physical and chemical properties govern whether a ligand will bind a protein. Oftentimes, binding is localized to a certain location on the protein, called the active site. The binding mechanism is analogous to encryption and decryption in some ways. Compounds can be thought of as encrypted files, where certain strings of atoms within the molecule are the strings of information hidden in the file. Proteins on the other hand, are analogous to decryption algorithms required for access to the encrypted data. When atoms of specific amino acids of a protein form bonds with atoms of the compound, the protein decodes the message being sent by the compound by altering its conformation (spatial alignment).
Though it is not inherent in all compound-protein interactions, conformational alteration of the protein is a common response to interaction with a compound. This detail is crucial for understanding that an odorant binding to the olfactory receptor adjusts conformation of the protein in a way that triggers a cascade of events. The change in conformation is necessary to ensure that the signal cascade is not continuously activated. The binding event is the “on-switch”.
Current methods of prediction for compound-ligand interactions are reliant on machine learning as well as extensive databases on compound and protein structures. Convolutional neural networks (CNNs) – like the MONN network architecture shown below – are wildly popular structures for protein-ligand modeling.
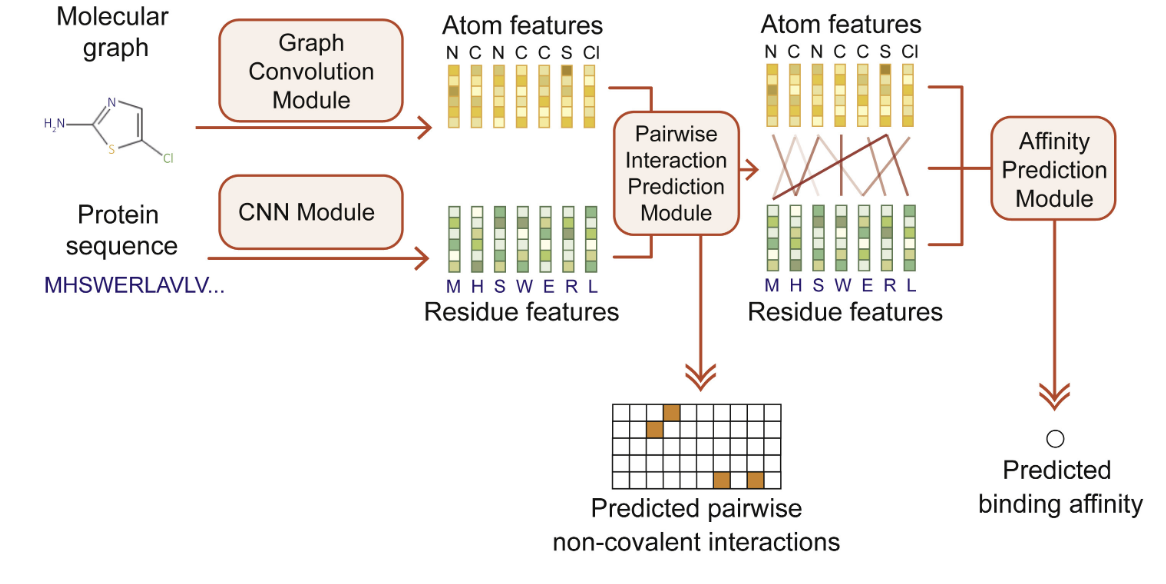
Other computational approaches – like mini-virtual screening – highlight the interactions between atoms of the compound and amino acids of the protein to isolate the active site for the interaction.
While we are able to visualize active sites and protein-compound interactions for proteins whose active sites have been successfully determined experimentally (frameworks like PyMol and Chimera are great visualization tools for protein-ligand interactions that have been experimentally confirmed), it is much more difficult to produce comparable visualizations for proteins whose structure is difficult to determine experimentally.
There is limited knowledge of the specific binding of olfactory receptor proteins to odorants because it is difficult to conduct experiments that accurately depict what is happening in vivo (in the body). Olfactory receptors are G-protein coupled receptors (GPCR). GPCRs have posed challenges for researchers as isolation of intact GPCRs through experimental means is almost impossible.
However, what we can deduce experimentally is whether or not a certain molecule will bind an olfactory receptor. In some cases, the binding affinity – how strong a compound’s interaction with the protein is – can be determined. Advances in machine learning, as well as the breakthroughs in protein modeling with Alpha Fold are promising steps toward a computational method for predicted compound-protein interactions in the near future.
A deep learning system that predicts a protein's 3D structure from its amino acid sequence, AlphaFold presents a viable – and experimentally validated – solution to the in vivo protein-folding problem. The AlphaFold project was started in 2016 by DeepMind, and the source code for the AI system was made available to the public in 2020. AlphaFold2, an updated version of AlphaFold has successfully achieved protein structure accuracy comparable to that of experimental procedures in labs. Generally, an accuracy of above 90% is considered to be within the threshold required for a predicted structure to be considered reliable. This bodes very well for researchers' ability to visualize and predict structures of GPCRs.
Amino acid sequences, the primary structure of proteins, serve as input to the program. The idea is to enter the amino acid sequence derived from the genetic code of a protein and receive a 3D model of the protein as output. The model consists of a recurrent convolutional neural network (CNN) applied to sequence alignments between the amino acid sequence of the input and amino acid sequences of homologous proteins with known 3D structures from genetic databases. Iterative refinement is implemented to continually adjust the conformation as the algorithm runs.
AlphaFold has revolutionized 3D protein modeling, but the system is simply the tip of the iceberg in terms of predicting the vast array of conformations individual proteins can take on. With promising results for structural analysis of GPCRs, depictions of olfactory receptors interacting with odorants is surely not far off.
Galaxolide is responsible for a musky scent humans are able to detect. AlphaFold has pretty high confidence in the model of this protein. Recent experiments revealed that galaxolide binds to the olfactory receptor encoded by the OR4D6 gene. A lack of this gene in humans results in the inability to detect the musky smell of galaxolide. The details of the binding of the two entities, which amino acid residues interact with which atoms of galaxolide, have yet to be discovered.
MDQINHTNVKEFFFLELTRSRELEFFLFVVFFAVYVATVLGNALIVVTITCESRLHTPMYFLLRNKSVLDIVFSSITVPKFLVDLLSDRKTISYNDCMAQIFFFHFAGGADIFFLSVMAYDRYLAIAKPLHYVTMMRKEVWVALVVASWVSGGLHSIIQVILMLPFPFCGPNTLDAFYCYVLQVVKLACTDTFALELFMISNNGLVTLLWFLLLLGSYTVILVMLRSHSGEGRNKALSTCTSHMLVVTLHFVPCVYIYCRPFMTLPMDTTISINNTVITPMLNPIIYSLRNQEMKSAMQRLQRRLGPSESRKWG
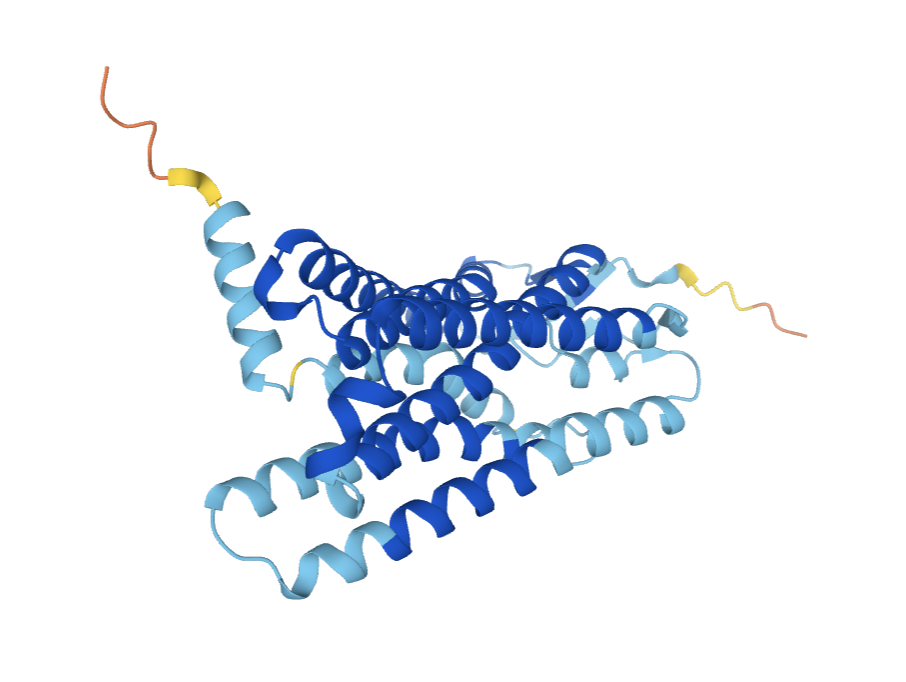
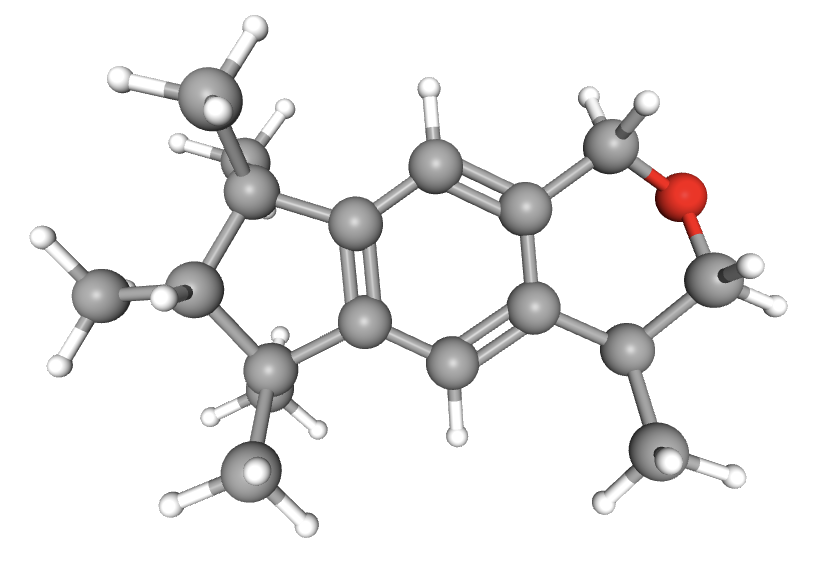
We chose this odorant-olfactory receptor combination to attempt to generate a 3D visualization of the interaction because it is a well-documented pairing. Further research into the area proved that identification of the active site has yet to be determined. Ultimately, we were unable to create such a visual due to a lack of resources and knowledge of generating an accurate prediction.
Olfactory receptors, like the majority of ligand binding proteins, are capable of binding to multiple odorants. OR4D6 is capable of attaching to a multitude of compounds, not just galaxolide. The converse is also true: galaxolide can be attached to a number of other olfactory receptors. They operate on the “many to many” rule!